AUSTIN, Texas —An international team of researchers, including Brett Baker from The University of Texas Marine Science Institute, has made the most comprehensive tree of life based on genomes, greatly expanding our view of the diversity of life on the planet. Using genetic data collected in recent years, the researchers found a group of bacteria that are so diverse genetically that they represent half of all the diversity of bacteria on the planet.
Much of this microbial diversity remained hidden until the genome revolution allowed microbiologists to search directly for new microbes in the environment by analyzing their genomes, rather than culturing them in a lab dish. Less than 0.1 percent of microbes in the world can be cultured, which means there are thousands, maybe even millions, of microbes yet to be discovered. According to Baker, the new technique of using genomes to discover new microorganisms is akin to the invention of the telescope and the proceeding discoveries of new stars and galaxies.
Until recently, most trees of life — which depict how life has evolved and diversified on the planet — were based on physical traits of organisms rather than their genes.
“The tree of life is our central organizing unit of biology,” said Assistant Professor Baker. “It’s the way we understand relationships amongst all life. Charles Darwin drew trees of what he was seeing in nature, and we now use genetic sequences to draw them.”
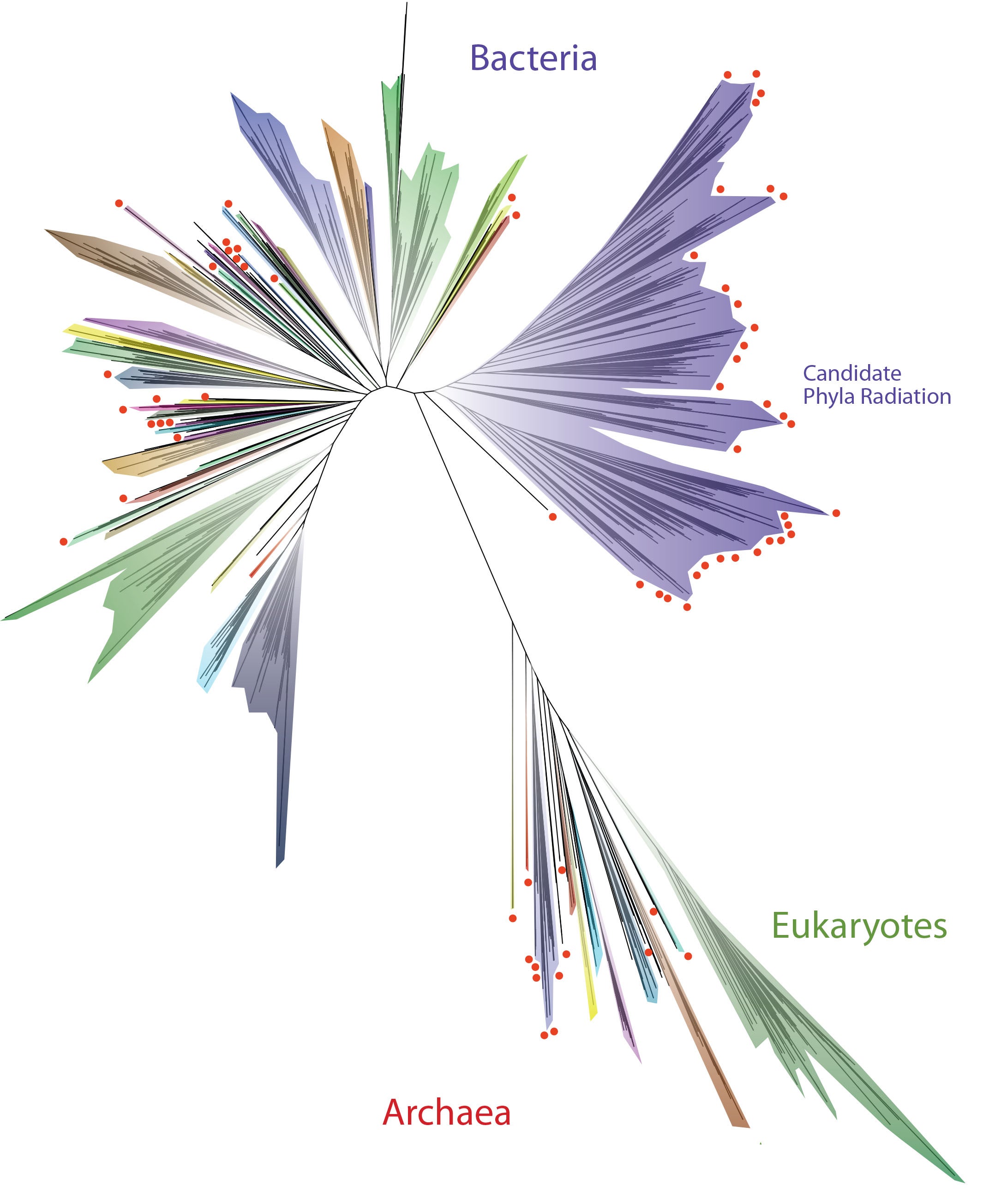
Scientists have created trees of life that include more species than this latest tree, but those tended to focus disproportionately on plants and animals. This newest tree — incorporating 3,083 organisms, one from each genus for which fully or almost fully sequenced genomes were available — more closely represents the full richness of life on the planet.
“This is the first three-domain genome-based tree to incorporate these uncultivable organisms, and it reveals the vast scope of as yet little known lineages,” said research team lead Jill Banfield from the University of California, Berkeley, referring to the three major domains of life: bacteria, Archaea and eukaryotes. “The new depiction will be of use not only to biologists who study microbial ecology, but also biochemists searching for novel genes and researchers studying evolution and Earth history.”
The new tree, published online April 11 in the journal Nature Microbiology, reinforces once again that the life we see around us — plants, animals, humans and other eukaryotes — represents a tiny percentage of the world’s biodiversity.
According to first author Laura Hug, the more than 1,000 newly reported organisms appearing on the revised tree are from a range of environments including a hot spring in Yellowstone National Park, a salt flat in Chile’s Atacama desert, terrestrial and wetland sediments, a sparkling water geyser, meadow soil and the inside of a dolphin’s mouth. All of these newly recognized organisms are known only from their genomes.
One striking aspect of the new tree of life is that a group of bacteria described as the “candidate phyla radiation” forms a very major branch. Only recognized recently and seemingly composed only of bacteria with symbiotic lifestyles, the candidate phyla radiation now appears to contain about half of all bacterial evolutionary diversity.
“This incredible diversity means that there are a mind boggling number of organisms that we are just beginning to explore the inner workings of, which could change our understanding of biology,” said Baker.
The paper’s authors are Laura Hug of the University of Waterloo; Jill Banfield, Karthik Anantharaman, Christopher Brown, Alexander Probst, Cindy Castelle, Cristina Butterfield, Brian Thomas, Alex Hernsdorf, Ronald Amundson and Kari Finstad of the University of California, Berkeley; Brett Baker from The University of Texas Marine Science Institute; Yuki Amano and Kotaro Ise of the Japan Atomic Energy Agency; Yohey Suzuki of the University of Tokyo; Natasha Dudek of the University of California, Santa Cruz; and David Relman of Stanford University.
The research was supported primarily by the Department of Energy (DOE) through Lawrence Berkeley National Laboratory, with metagenomic sequencing by DOE’s Joint Genome Institute in Walnut Creek, California.
High-res. image of the new tree of life available at Flickr.
This press release is based on a release by The University of California, Berkeley.



