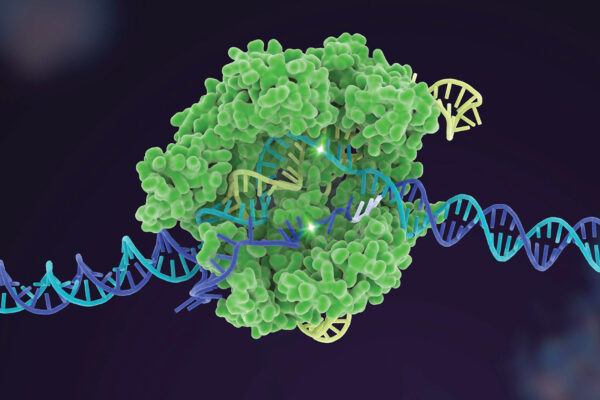
AUSTIN, Texas — One of the biggest scientific advances of the last decade is getting better thanks to researchers at The University of Texas at Austin; the University of California, Berkeley; and Korea University. The team has developed a new tool to help scientists choose the best available gene-editing option for a given job, making the technology called CRISPR safer, cheaper and more efficient. The tool is outlined in a paper out today in Nature Biotechnology.
The CRISPR gene-editing technique holds tremendous potential to improve human health, agriculture and the future of people on the planet, but the challenge lies in the delicate nature of gene editing — there is almost no room for error.
To edit genes, scientists use dozens of different enzymes from a naturally occurring system called CRISPR. Researchers locate a problematic DNA sequence and use these specialized enzymes to snip it as if using a pair of scissors, allowing genetic material to be added, removed or altered. But these scissors are not perfect. Accuracy and effectiveness vary by the CRISPR enzyme and the project. The new tool guides users, so they can pick the best CRISPR enzyme for their high-stakes gene edit.
“We designed a new method that tests the specificity of these different CRISPR enzymes — how precise they are — robustly against any changes to the DNA sequence that could misdirect them, and in a cleaner way than has ever been done before,” said Steve Jones, a UT research scientist who co-wrote the paper with Ilya Finkelstein, an associate professor of molecular biosciences.
Problems can occur when a CRISPR enzyme targets the wrong sections of DNA. Each CRISPR enzyme has strengths and weaknesses in editing different sequences, so the researchers set out to create a tool to help scientists compare the different enzymes and find the best one for a given job.
“CRISPR wasn’t designed in a lab. It wasn’t made by humans for humans. It was made by bacteria to defend against viruses,” said John Hawkins, a Ph.D. alumnus who was recently with UT’s Oden Institute for Computational Engineering and Sciences. “There is incredible potential for its use in medicine, but the first rule of medicine is ‘do no harm.’ Our work is trying to make CRISPR safer.”
The team of researchers developed a library of DNA sequences and measured how accurate each CRISPR enzyme was, how long it took the enzyme to edit the sequences and how precisely they edited the sequence. For some tasks the commonly used enzyme CRISPR-Cas9 worked best; in others, different enzymes performed much better.
“It’s like a standardized test,” Hawkins said. “Every student gets the same test, and now you have a benchmark to compare them.”
The tool allows scientists to choose the best enzyme for editing on the first try, so the process becomes more efficient and cheaper. Additionally, it gives scientists information about where mistakes are most likely to occur for each enzyme, saving time.
“This technique gives us a new way to reduce risk,” Jones said. “It allows gene edits to be more predictable.”
Nicole V. Johnson, Kuang Hu, James R. Rybarski, William H. Press and Ilya J. Finkelstein of The University of Texas at Austin; Cheulhee Jung of Korea University; and Janice S. Chen and Jennifer A. Doudna of University of California, Berkeley, contributed to the research.
The research was funded by a College of Natural Sciences Catalyst Grant, The Welch Foundation and the National Institutes of Health.